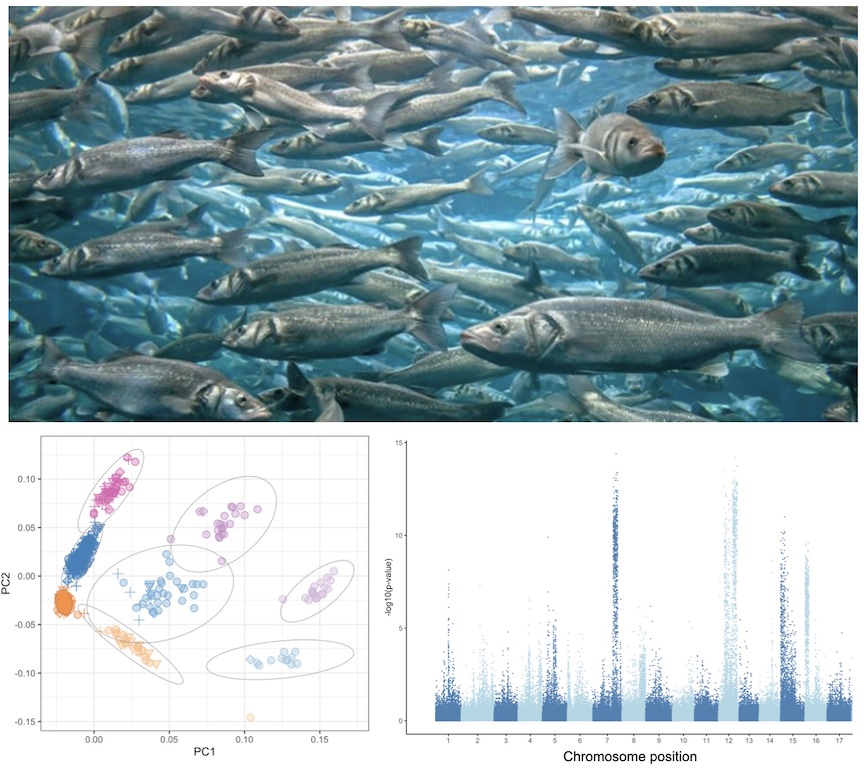
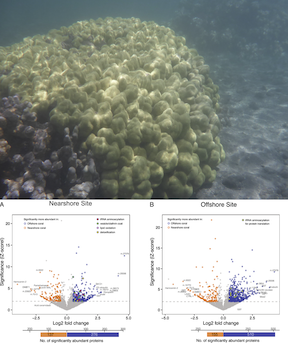
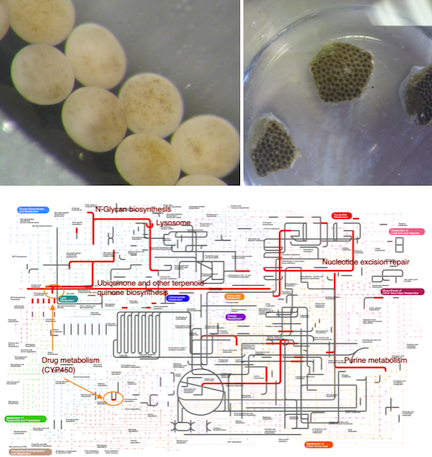
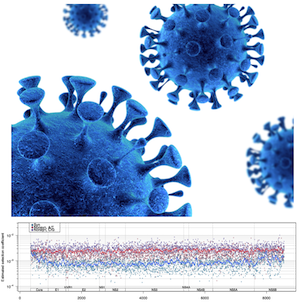
Marine Biologist, Molecular Ecologist, Computational Biologist

My research interests are focused on understanding how marine organisms respond to environmental stressors and adapt to changing environments. Anthropogenic stressors have changed our coastal marine environments drastically over the last century. I am interested in understanding what makes some individuals, populations, or species more resilient than others to individual or combinations of stressors. I approach these questions using controlled laboratory experiments, field experiments, and field surveys together with molecular 'omics' techniques, such as genomics and proteomics. I use an ecotoxicological approach to understand physiological and molecular responses of organisms to specific stressors, and employ genomics and proteomics combined with ecological experiments to answer ecological and evolutionary processes behind local adaptation, acclimatization, and resilience in marine organisms. The ultimate goal of my research is to provide solid scientific data that will lead to the protection and conservation of marine biodiversity. My primary interest is with coral reef ecosystems, and specifically, I have worked extensively with reef-building corals. I have also worked with other marine organisms, including sea urchins, nudibranch, and algae. I am interested in working with different marine systems as opportunities present. I am also passionate about computational biology and promoting coding for STEM/biology students.